Copy phyloseq otu_table data to sample_data
Arguments
- ps
phyloseq with sample_data
- taxa
list of taxa_names to copy to sample_data, or NULL (which selects all with
phyloseq::taxa_names())
Examples
library(phyloseq)
data("dietswap", package = "microbiome")
ps <- dietswap %>% ps_otu2samdat("Akkermansia")
sample_variables(ps)
#> [1] "subject" "sex" "nationality"
#> [4] "group" "sample" "timepoint"
#> [7] "timepoint.within.group" "bmi_group" "Akkermansia"
# or if you do not specify any taxa, all are copied
ps_all <- dietswap %>% ps_otu2samdat()
sample_variables(ps_all)[1:15]
#> [1] "subject" "sex"
#> [3] "nationality" "group"
#> [5] "sample" "timepoint"
#> [7] "timepoint.within.group" "bmi_group"
#> [9] "Actinomycetaceae" "Aerococcus"
#> [11] "Aeromonas" "Akkermansia"
#> [13] "Alcaligenes faecalis et rel." "Allistipes et rel."
#> [15] "Anaerobiospirillum"
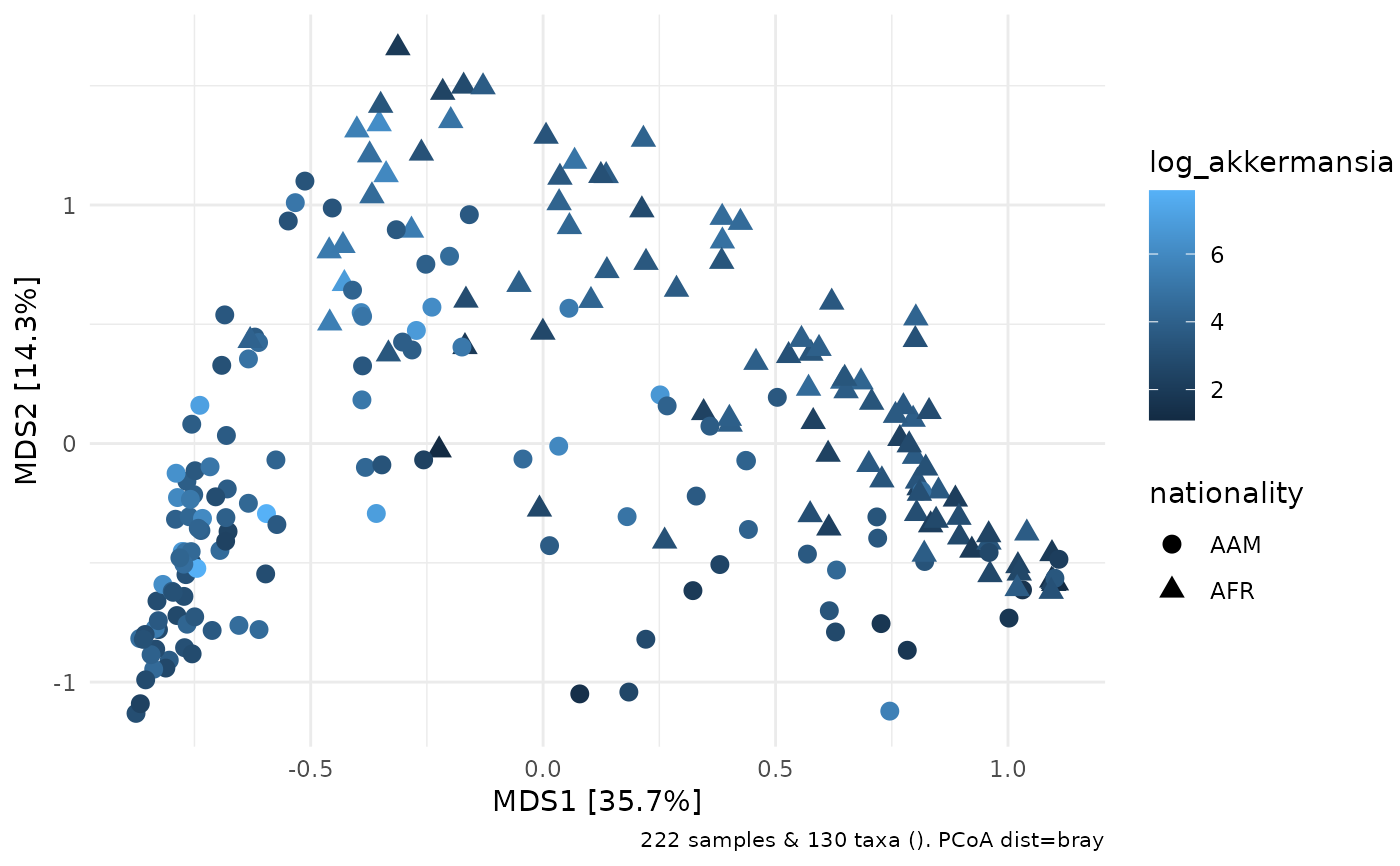
# this could be useful for colouring ordination plots, for example
ps %>%
ps_mutate(log_akkermansia = log(Akkermansia)) %>%
dist_calc("bray") %>%
ord_calc(method = "PCoA") %>%
ord_plot(
colour = "log_akkermansia",
size = 3, shape = "nationality"
)