
Sort phyloseq samples by ordination axes scores
Source:R/ps_sort_ord.R
ordination-sorting-samples.Rdps_sort_ord reorders samples in a phyloseq object based on their relative
position on 1 or 2 ordination axes.
ord_order_samples gets the sample_names in order from the ordination
contained in a psExtra. This is used internally by ps_sort_ord
If 2 axes given, the samples are sorted by anticlockwise rotation around the selected ordination axes, starting on first axis given, upper right quadrant. (This is used by ord_plot_iris.)
If 1 axis is given, samples are sorted by increasing value order along this axis. This could be used to arrange samples on a rectangular barplot in order of appearance along a parallel axis of a paired ordination plot.
Arguments
- ps
phyloseq object to be sorted
- ord
psExtra with ordination object
- axes
which axes to use for sorting? numerical vector of length 1 or 2
- scaling
Type 2, or type 1 scaling. For more info, see https://sites.google.com/site/mb3gustame/constrained-analyses/redundancy-analysis. Either "species" or "site" scores are scaled by (proportional) eigenvalues, and the other set of scores is left unscaled (from ?vegan::scores.cca)
See also
These functions were created to support ordering of samples on
ord_plot_iristax_sort_ordfor ordering taxa in phyloseq by ordination
Examples
# attach other necessary packages
library(ggplot2)
# example data
ibd <- microViz::ibd %>%
tax_filter(min_prevalence = 2) %>%
tax_fix() %>%
phyloseq_validate()
# create numeric variables for constrained ordination
ibd <- ibd %>%
ps_mutate(
ibd = as.numeric(ibd == "ibd"),
steroids = as.numeric(steroids == "steroids"),
abx = as.numeric(abx == "abx"),
female = as.numeric(gender == "female"),
# and make a shorter ID variable
id = stringr::str_remove_all(sample, "^[0]{1,2}|-[A-Z]+")
)
# create an ordination
ordi <- ibd %>%
tax_transform("clr", rank = "Genus") %>%
ord_calc()
ord_order_samples(ordi, axes = 1) %>% head(8)
#> [1] "012A" "031A" "025A" "199A" "030A" "041A" "048A" "119A"
ps_sort_ord(ibd, ordi, axes = 1) %>%
phyloseq::sample_names() %>%
head(8)
#> [1] "012A" "031A" "025A" "199A" "030A" "041A" "048A" "119A"
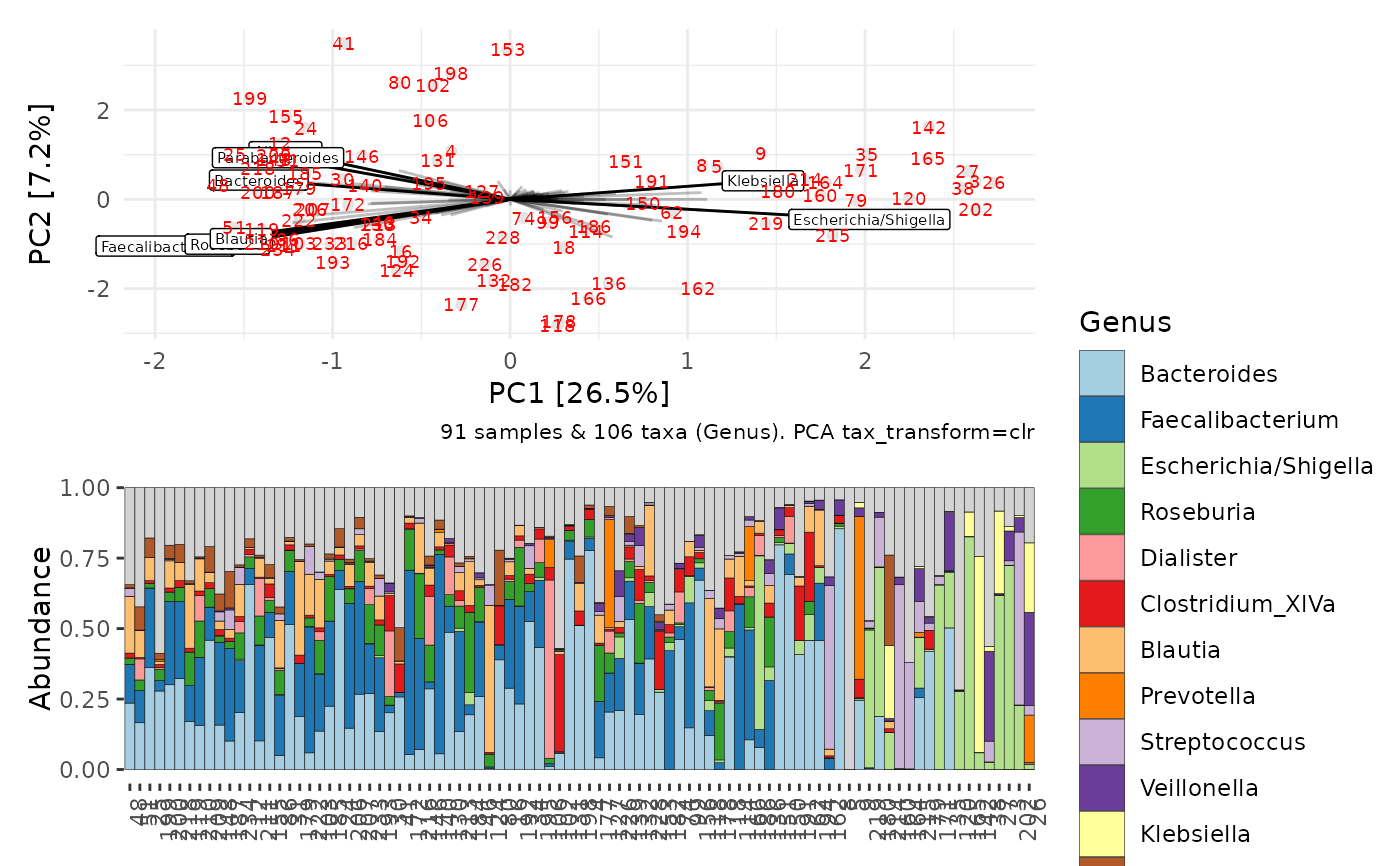
p1 <- ord_plot(ordi, colour = "grey90", plot_taxa = 1:8, tax_vec_length = 1) +
geom_text(aes(label = id), size = 2.5, colour = "red")
b1 <- ibd %>%
ps_sort_ord(ord = ordi, axes = 1) %>%
comp_barplot(
tax_level = "Genus", n_taxa = 12, label = "id",
order_taxa = ord_order_taxa(ordi, axes = 1),
sample_order = "asis"
) +
theme(axis.text.x = element_text(angle = 90, hjust = 1))
patchwork::wrap_plots(p1, b1, ncol = 1)
 # constrained ordination example (and match vertical axis) #
cordi <- ibd %>%
tax_transform("clr", rank = "Genus") %>%
ord_calc(
constraints = c("steroids", "abx", "ibd"), conditions = "female",
scale_cc = FALSE
)
cordi %>% ord_plot(plot_taxa = 1:6, axes = 2:1)
# constrained ordination example (and match vertical axis) #
cordi <- ibd %>%
tax_transform("clr", rank = "Genus") %>%
ord_calc(
constraints = c("steroids", "abx", "ibd"), conditions = "female",
scale_cc = FALSE
)
cordi %>% ord_plot(plot_taxa = 1:6, axes = 2:1)
