Helper to specify a HeatmapAnnotation for samples in comp_heatmap
Source:R/heatmaps-sampleAnnotation.R
sampleAnnotation.RdHelper to specify a HeatmapAnnotation for samples in comp_heatmap
Usage
sampleAnnotation(
...,
name,
annotation_legend_param = list(),
show_legend = TRUE,
gp = grid::gpar(col = NA),
border = FALSE,
gap = grid::unit(2, "mm"),
show_annotation_name = TRUE,
annotation_label = NULL,
annotation_name_gp = grid::gpar(),
annotation_name_offset = NULL,
annotation_name_rot = NULL,
annotation_name_align = FALSE,
annotation_name_side = "auto",
.data = NULL,
.samples = NULL,
.side = NULL
)Arguments
- ...
Name-value pairs where the names correspond to annotation names and values are the output of sample annotation functions such as anno_sample(), or manually specified AnnotationFunction objects
- name
Name of the heatmap annotation, optional.
- annotation_legend_param
A list which contains parameters for annotation legends. See
color_mapping_legend,ColorMapping-methodfor all possible options.- show_legend
Whether show annotation legends. The value can be one single value or a vector.
- gp
Graphic parameters for simple annotations (with
fillparameter ignored).- border
border of single annotations.
- gap
Gap between annotations. It can be a single value or a vector of
unitobjects.- show_annotation_name
Whether show annotation names? For column annotation, annotation names are drawn either on the left or the right, and for row annotations, names are draw either on top or at the bottom. The value can be a vector.
- annotation_label
Labels for the annotations. By default it is the same as individual annotation names.
- annotation_name_gp
Graphic parameters for annotation names. Graphic parameters can be vectors.
- annotation_name_offset
Offset to the annotation names, a
unitobject. The value can be a vector.- annotation_name_rot
Rotation of the annotation names. The value can be a vector.
- annotation_name_align
Whether to align the annotation names.
- annotation_name_side
Side of the annotation names.
- .data
OPTIONAL phyloseq or psExtra, only set this to override use of same data as in heatmap
- .samples
OPTIONAL selection vector of sample names, only set this if providing .data argument to override default
- .side
OPTIONAL string, indicating the side for the variable annotations: only set this to override default
Examples
library("ComplexHeatmap")
data("ibd", package = "microViz")
psq <- tax_filter(ibd, min_prevalence = 5)
psq <- tax_mutate(psq, Species = NULL)
psq <- tax_fix(psq)
psq <- tax_agg(psq, rank = "Family")
taxa <- tax_top(psq, n = 15, rank = "Family")
samples <- phyloseq::sample_names(psq)
set.seed(42) # random colours used in first example
# sampleAnnotation returns a function that takes data, samples, and which
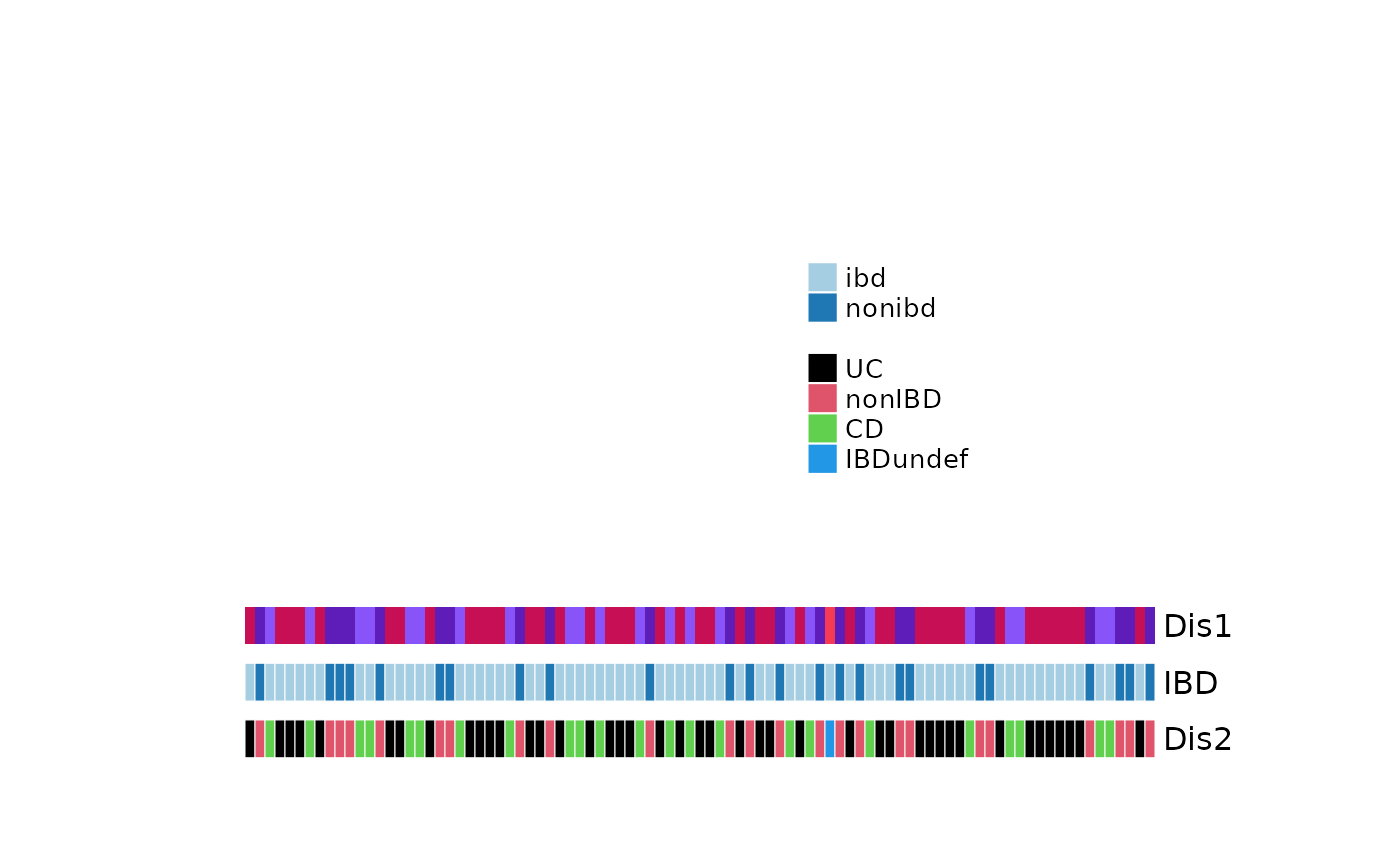
fun <- sampleAnnotation(
gap = grid::unit(2.5, "mm"),
Dis1 = anno_sample(var = "DiseaseState"),
IBD = anno_sample_cat(var = "ibd"),
Dis2 = anno_sample_cat(var = "DiseaseState", col = 1:4)
)
# manually specify the sample annotation function by giving it data etc.
heatmapAnnoFunction <- fun(.data = psq, .side = "top", .samples = samples)
# draw the annotation without a heatmap, you will never normally do this!
grid.newpage()
vp <- viewport(width = 0.65, height = 0.75)
pushViewport(vp)
draw(heatmapAnnoFunction)
pushViewport(viewport(x = 0.7, y = 0.6))
draw(attr(heatmapAnnoFunction, "Legends"))