
Make new phyloseq taxa names from classification and taxon abundance info
Source:R/tax_rename.R
tax_rename.RdPairs classification at the given rank with a numeric ranking suffix (based on abundance or prevalence data) to automatically create informative taxa names.
Usage
tax_rename(
ps,
rank,
sort_by = sum,
transform_for_sort = "identity",
pad_digits = "auto",
sep = " ",
...
)Arguments
- ps
phyloseq object
- rank
name of rank to use in new taxa names
- sort_by
how to sort taxa for numbering within rank-based groups (a tax_sort option)
- transform_for_sort
named of transformation to apply to taxa before sorting
- pad_digits
how long should the numeric suffixes be? see details
- sep
character to separate the rank prefixes from numeric suffixes
- ...
additional arguments passed to tax_sort
Details
e.g. "Bacteroides 003" for the third most abundant Bacteroides OTU or ASV.
Taxa are returned in original order, and otu_table is returned un-transformed.
pad_digits options:
"auto" –> minimum digits to have equal length numbers within groups
"max" –> minimum digits to have equal length numbers across all groups
A number: e.g.
3 –> 001, 002, ..., 042, ..., 180, ...
1 –> 1, 2, ..., 42, ..., 180, ...
See also
phyloseq::taxa_names for accessing and manually setting names
Examples
library(phyloseq)
data("ibd", package = "microViz")
ps <- ibd %>%
tax_filter(min_prevalence = 3) %>%
tax_fix()
# show a few of the current, uninformative names
taxa_names(ps) %>% head(15)
#> [1] "OTU.49" "OTU.50" "OTU.52" "OTU.54" "OTU.56" "OTU.57" "OTU.59"
#> [8] "OTU.76" "OTU.90" "OTU.92" "OTU.108" "OTU.142" "OTU.185" "OTU.197"
#> [15] "OTU.200"
taxa_names(ps) %>% tail(15)
#> [1] "OTU.36191" "OTU.36241" "OTU.36243" "OTU.36244" "OTU.36245" "OTU.36247"
#> [7] "OTU.36248" "OTU.36290" "OTU.36292" "OTU.36293" "OTU.36294" "OTU.36295"
#> [13] "OTU.36297" "OTU.36300" "OTU.36301"
# change names to genus classification plus number
psNewNames <- ps %>% tax_rename(rank = "Genus")
taxa_names(psNewNames) %>% head(15)
#> [1] "Faecalibacterium 203" "Escherichia/Shigella 081"
#> [3] "Clostridium_XlVa 045" "Lachnospiraceae Family 20"
#> [5] "Bacteroides 0825" "Flavonifractor 25"
#> [7] "Ruminococcaceae Family 05" "Clostridium_XlVa 028"
#> [9] "Faecalibacterium 204" "Escherichia/Shigella 088"
#> [11] "Escherichia/Shigella 130" "Clostridium_XlVa 110"
#> [13] "Lachnospiraceae Family 58" "Lachnospiracea_incertae_sedis 34"
#> [15] "Bacteroides 0967"
taxa_names(psNewNames) %>% tail(15)
#> [1] "Oscillibacter 22" "Dialister 067"
#> [3] "Bifidobacterium 36" "Roseburia 123"
#> [5] "Oscillibacter 19" "Lachnospiracea_incertae_sedis 45"
#> [7] "Enterococcus 15" "Bacteroides 0205"
#> [9] "Bacteroides 0187" "Bacteroides 0165"
#> [11] "Dialister 015" "Bacteroides 0142"
#> [13] "Bacteroides 0338" "Dorea 04"
#> [15] "Bacteroides 0226"
# demonstrate some alternative argument settings
psNewNames2 <- ps %>% tax_rename(
rank = "Family", sort_by = prev, pad_digits = "max", sep = "-"
)
taxa_names(psNewNames2) %>% head(15)
#> [1] "Ruminococcaceae-0216" "Enterobacteriaceae-0101"
#> [3] "Lachnospiraceae-0191" "Lachnospiraceae-0192"
#> [5] "Bacteroidaceae-0755" "Ruminococcaceae-0217"
#> [7] "Ruminococcaceae-0218" "Lachnospiraceae-0082"
#> [9] "Ruminococcaceae-0361" "Enterobacteriaceae-0140"
#> [11] "Enterobacteriaceae-0141" "Lachnospiraceae-0404"
#> [13] "Lachnospiraceae-0405" "Lachnospiraceae-0317"
#> [15] "Bacteroidaceae-0756"
taxa_names(psNewNames2) %>% tail(15)
#> [1] "Ruminococcaceae-0522" "Veillonellaceae-0223"
#> [3] "Bifidobacteriaceae-0036" "Lachnospiraceae-0623"
#> [5] "Ruminococcaceae-0523" "Lachnospiraceae-0403"
#> [7] "Enterococcaceae-0018" "Bacteroidaceae-0207"
#> [9] "Bacteroidaceae-0280" "Bacteroidaceae-0754"
#> [11] "Veillonellaceae-0130" "Bacteroidaceae-0143"
#> [13] "Bacteroidaceae-0426" "Lachnospiraceae-0036"
#> [15] "Bacteroidaceae-0208"
ps %>%
tax_rename(rank = "Genus", pad_digits = 2) %>%
taxa_names() %>%
head(15)
#> [1] "Faecalibacterium 203" "Escherichia/Shigella 81"
#> [3] "Clostridium_XlVa 45" "Lachnospiraceae Family 20"
#> [5] "Bacteroides 825" "Flavonifractor 25"
#> [7] "Ruminococcaceae Family 05" "Clostridium_XlVa 28"
#> [9] "Faecalibacterium 204" "Escherichia/Shigella 88"
#> [11] "Escherichia/Shigella 130" "Clostridium_XlVa 110"
#> [13] "Lachnospiraceae Family 58" "Lachnospiracea_incertae_sedis 34"
#> [15] "Bacteroides 967"
# naming improvement on plots example
library(ggplot2)
library(patchwork)
# Overly aggressive OTU filtering to simplify and speed up example
psExample <- ps %>% tax_filter(min_prevalence = 0.4)
#> Proportional min_prevalence given: 0.4 --> min 37/91 samples.
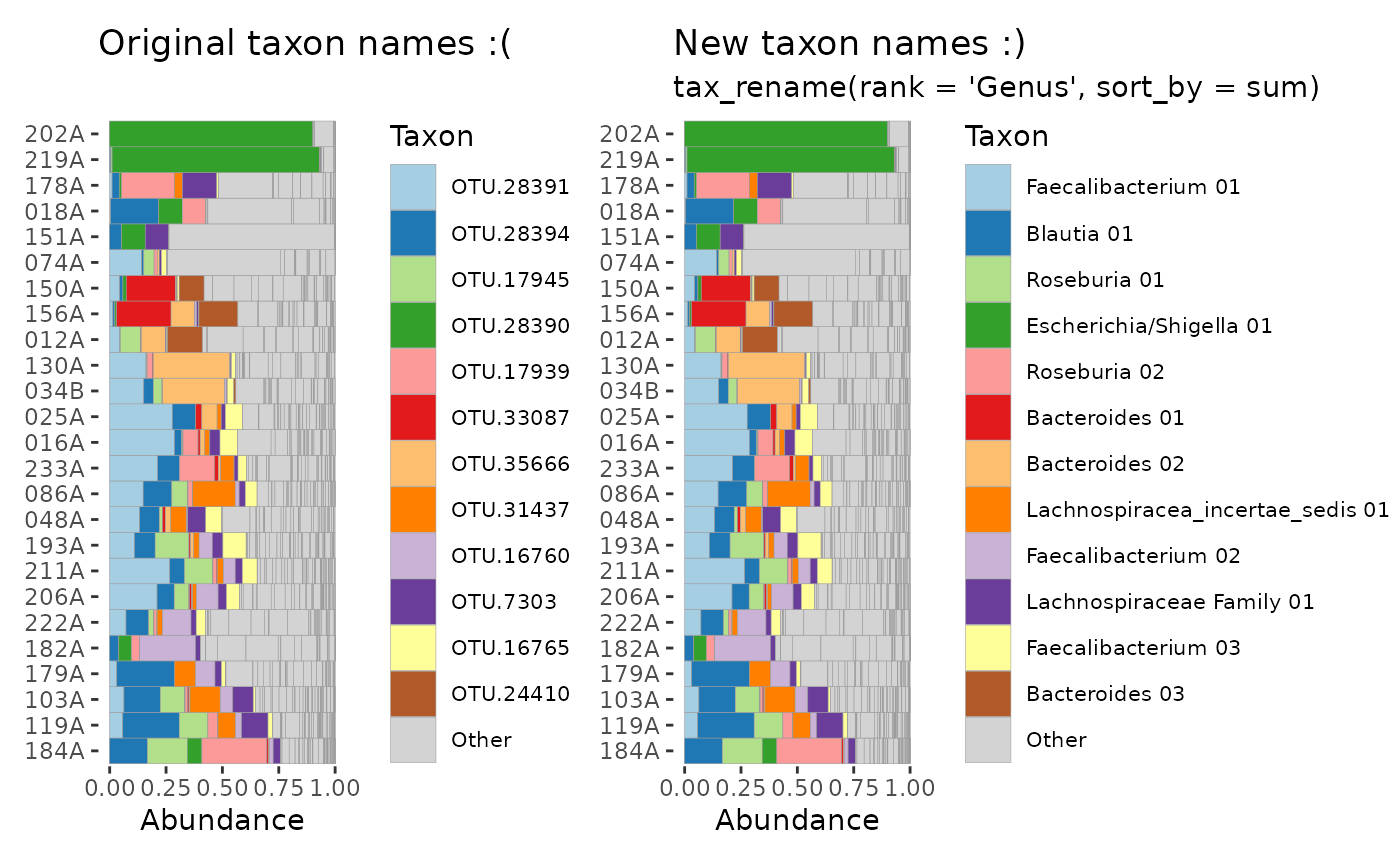
# before OTU renaming
before <- psExample %>%
ps_filter(activity == "inactive") %>%
tax_names2rank("Taxon") %>%
comp_barplot(
tax_level = "Taxon", n_taxa = 12, other_name = "Other",
merge_other = FALSE, bar_outline_colour = "grey60"
) +
coord_flip() +
ggtitle("Original taxon names :(")
# after OTU renaming
after <- psExample %>%
ps_filter(activity == "inactive") %>%
tax_rename(rank = "Genus", pad_digits = "max") %>%
tax_names2rank("Taxon") %>%
comp_barplot(
tax_level = "Taxon", n_taxa = 12, other_name = "Other",
merge_other = FALSE, bar_outline_colour = "grey60"
) +
coord_flip() +
ggtitle("New taxon names :)", "tax_rename(rank = 'Genus', sort_by = sum)")
before + after & theme(legend.text = element_text(size = 8))
 # ordination example
psExample %>%
tax_rename(rank = "Genus", sort_by = sum) %>%
tax_names2rank("otu") %>%
tax_transform("clr", rank = "otu") %>%
ord_calc() %>%
ord_plot(
size = 2, colour = "ibd", shape = "circle", alpha = 0.5,
plot_taxa = 1:10,
tax_vec_length = 0.5,
tax_lab_style = tax_lab_style(
type = "text", max_angle = 90, check_overlap = TRUE,
size = 2.5, fontface = "bold"
),
tax_vec_style_all = vec_tax_all(alpha = 0.1)
) +
coord_fixed(clip = "off") +
stat_chull(aes(colour = ibd)) +
scale_colour_brewer(palette = "Dark2") +
theme(panel.grid = element_line(size = 0.1))
#> Warning: The `size` argument of `element_line()` is deprecated as of ggplot2 3.4.0.
#> ℹ Please use the `linewidth` argument instead.
# ordination example
psExample %>%
tax_rename(rank = "Genus", sort_by = sum) %>%
tax_names2rank("otu") %>%
tax_transform("clr", rank = "otu") %>%
ord_calc() %>%
ord_plot(
size = 2, colour = "ibd", shape = "circle", alpha = 0.5,
plot_taxa = 1:10,
tax_vec_length = 0.5,
tax_lab_style = tax_lab_style(
type = "text", max_angle = 90, check_overlap = TRUE,
size = 2.5, fontface = "bold"
),
tax_vec_style_all = vec_tax_all(alpha = 0.1)
) +
coord_fixed(clip = "off") +
stat_chull(aes(colour = ibd)) +
scale_colour_brewer(palette = "Dark2") +
theme(panel.grid = element_line(size = 0.1))
#> Warning: The `size` argument of `element_line()` is deprecated as of ggplot2 3.4.0.
#> ℹ Please use the `linewidth` argument instead.
